Diffusion-based Personalized Pathology Disentanglement for Impaired Gait Analysis
AAAI 2026
Abstract
In the context of global population aging, the prevalence of neurodegenerative diseases is rapidly increasing. Vision-based impaired gait analysis emerges as a promising alternative for automatic and non-invasive diagnosis. While prior efforts have advanced either accuracy or interpretability of gait analysis, few have effectively addressed both aspects in a unified framework. To bridge this gap, we propose DPPD, a Diffusion-based Personalized Pathology Disentanglement model that jointly performs quantitative gait scoring, dementia subtyping, and qualitative anomaly highlighting. Motivated by the observation that pathological gait features exhibit stronger inter-class separability across different gait severity than raw features, DPPD is proposed based on the subject-specific pathology disentanglement perspective. Specifically, it comprises three key components: (1) a 3DmotionBERT for encoding gait representation from 3D human pose sequences estimated, (2) a latent diffusion-based Gait Denoiser for generating personalized normal gait features, and (3) a Dual Pathology Disentanglement mechanism that captures both static pose and dynamic motion pathological representation from the residual between raw and normal gait features. These disentangled pathologies further enable quantitative classification and qualitative anomaly highlighting. Experiments on the PDGait and 3DGait datasets demonstrate that DPPD outperforms state-of-the-art methods in classification accuracy while providing reliable and interpretable visualizations of gait anomalies.
Overview
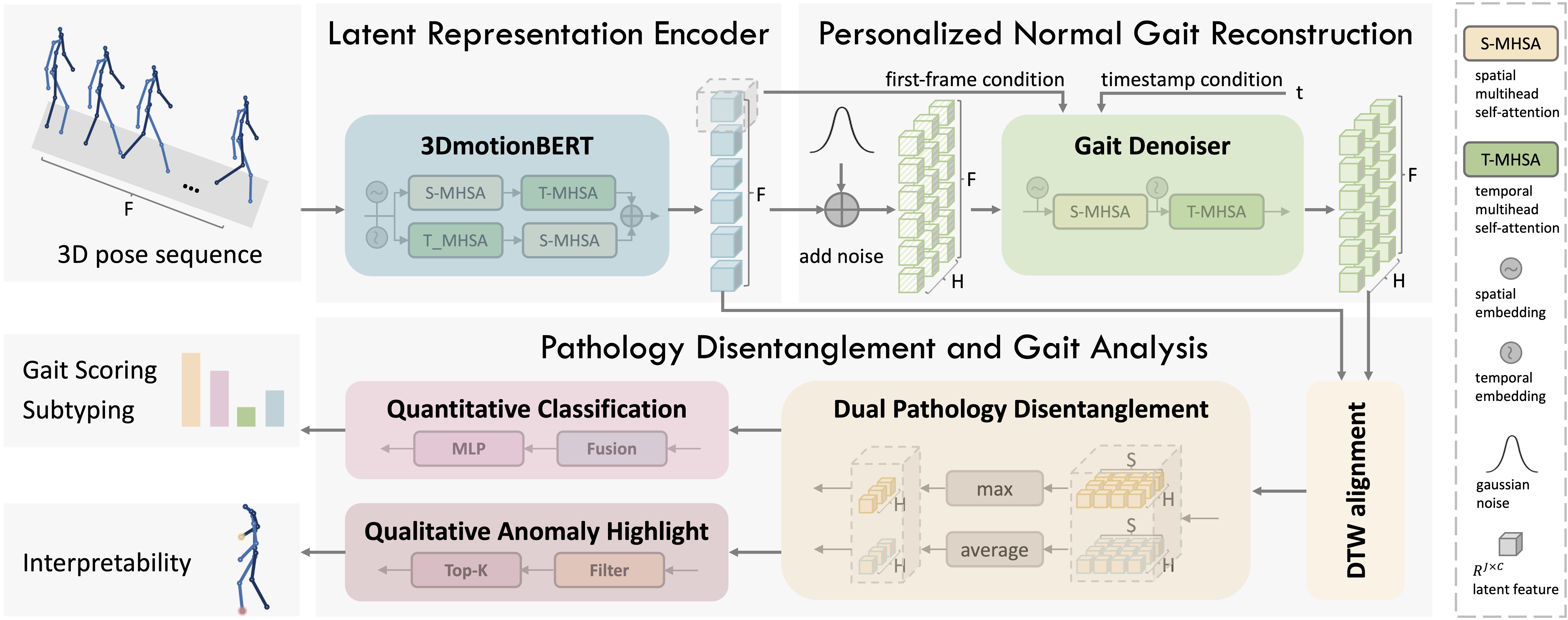
A 3D pose sequence is first encoded into a latent representation using 3DmotionBERT. The GD module then generates the corresponding normal gaits from a noisy version of the encoded input. Based on the original and generated normal representations, dual pathological features are extracted through DPD and used for both quantitative gait classification and qualitative anomaly highlighting.
Highlights
1) Pathology Disentanglement Framework.
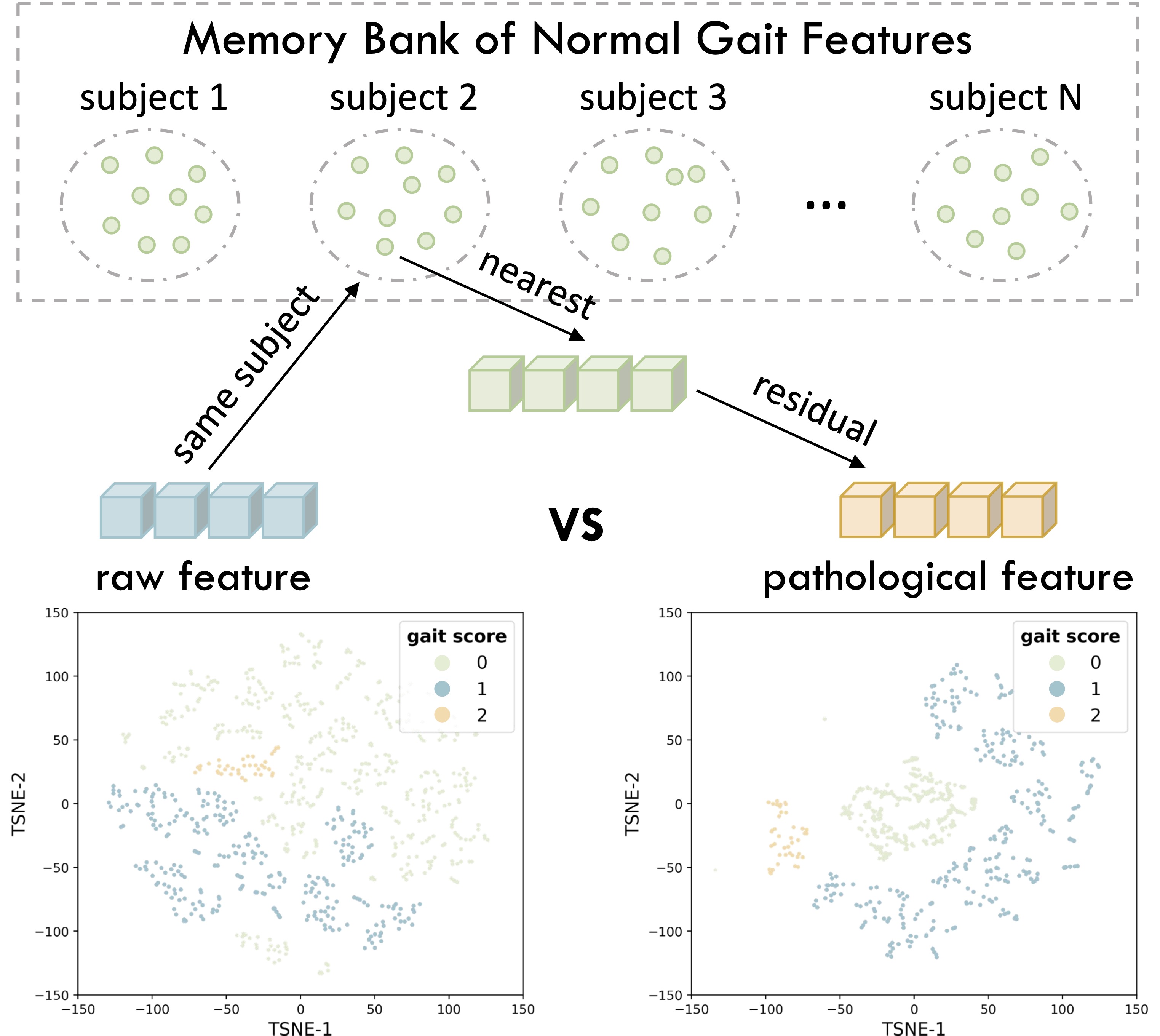
As shown by the comparison between residual and raw gait features, the residual features demonstrate superior inter-class separability across different severity levels, indicating that explicitly disentangling pathological components can enhance gait analysis. To this end, we propose DPPD, a novel framework for impaired gait analysis that, for the first time, explicitly disentangles pathological features, thereby enabling both accurate and interpretable gait assessment for neurodegenerative diseases (NDDs).
2) Personalized Normal Gait Reconstruction.
To reconstruct the normal gait representation of a patient, we propose a latent diffusion model, termed GD, which learns the distribution of normal gait representations in the latent space. Notably, normal gait patterns vary across individuals due to differences in skeletal structure and motion habits. To account for such variability, we introduce two complementary techniques, First-frame Condition and Personality Contrastive Loss, which embed subject-specific characteristics to generate personalized normal gait representations.
3) Dual Pathology Disentanglement.
We disentangle pathological features in each hypothesis via residual computation. Inspired by prior works such as FSGait and Dual-Conditioned Motion Diffusion, which suggest that motion anomalies can manifest as both static pose and dynamic motion deviations, we design a DPD module to capture these two pathological patterns.
Experiments
Gait Scoring
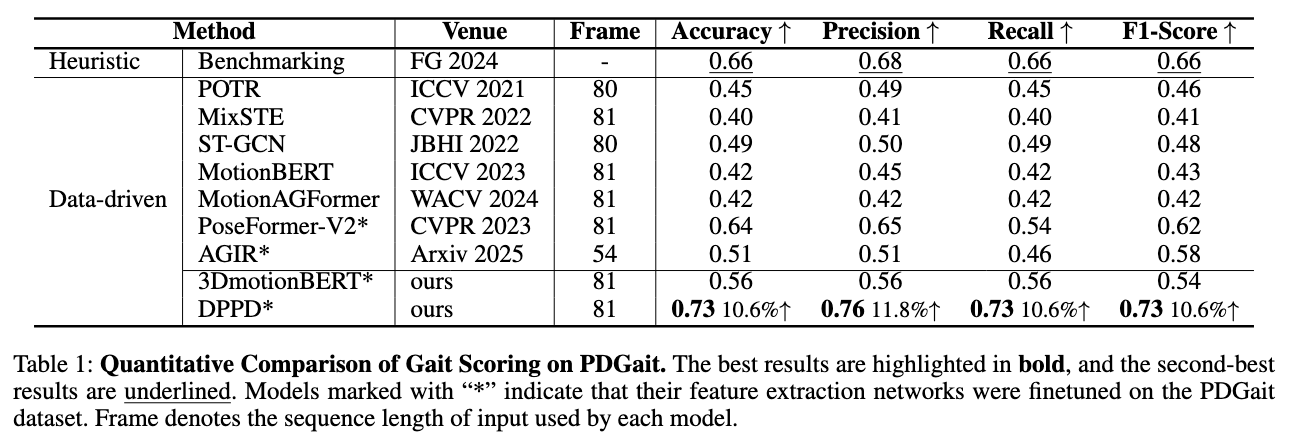
Experiments demonstrate that, compared to existing heuristic methods and data-driven methods, our proposed DPPD shows an accuracy advantage in gait anomaly scoring tasks on the PDGait clinical gait dataset.
Disease Subtyping
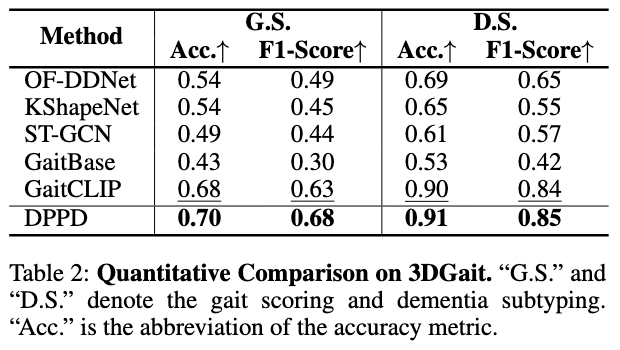
Experiments demonstrate that, compared to existing methods, our proposed DPPD shows superior accuracy in both gait scoring and disease subtyping tasks on the 3DGait clinical gait dataset, proving the generalizability of DPPD across multiple tasks.
Qualitative Anomaly Highlight
The visualization results demonstrate the capbility to reconstruct normal gait features and highlight the abnormal joints of the proposed DPPD.

Citation
@inproceedings{wan2026diffusion,
title = {Diffusion-based Personalized Pathology Disentanglement for Impaired Gait Analysis},
author = {Wan, Xiaoyue and Zhao, Xu},
booktitle = {Proceedings of the AAAI Conference on Artificial Intelligence},
year = {2026},
}
